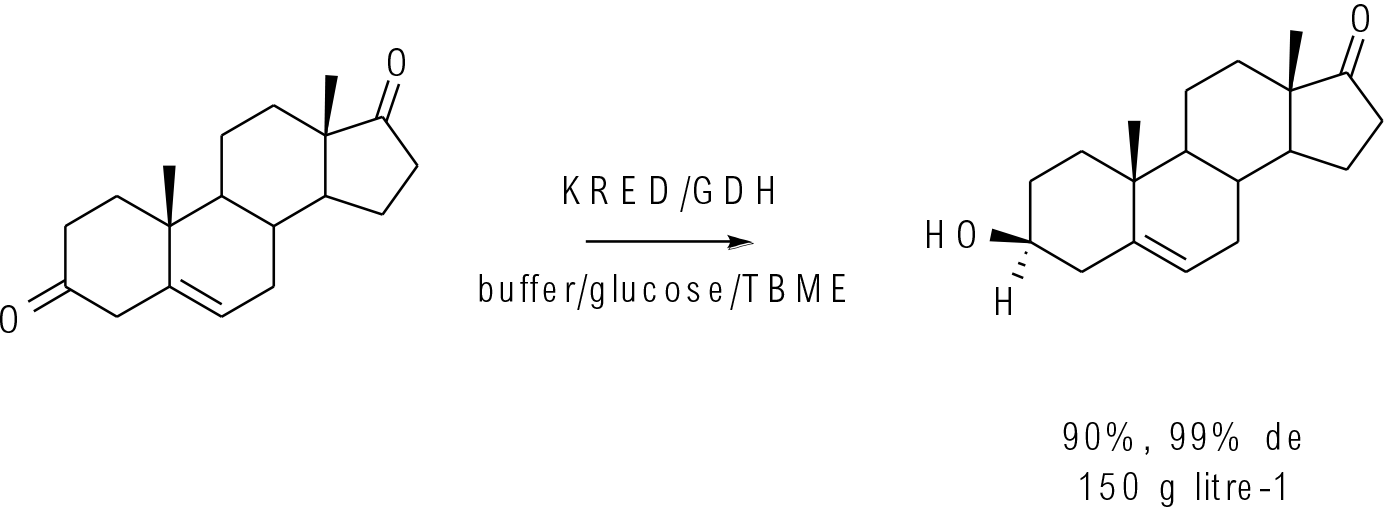Mechanism + Description
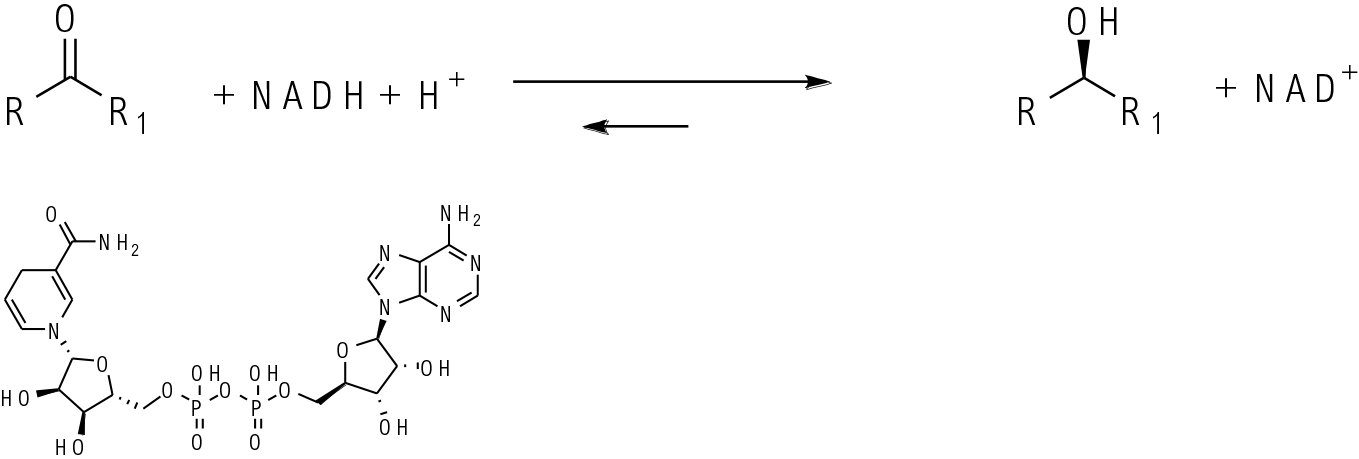
KREDs contain a metal- Zn2+ at the active site plus a cofactor NAD(P)H. The ketone binds in the enzyme active site and a hydride is delivered to one prochiral face of the ketone from the cofactor. The oxidised cofactor then needs to be reduced back to complete the catalytic cycle.
General comments
A wide range of ADH/KRED enzymes now exist with both R and S selective enzymes available. There are now many examples of ADH/KRED enzymes being evolved to reduce more complex/hindered ketone substrates. Key to the success of a ADH/KRED reduction is driving the NAD(P)H cofactor recycle. This is normally done using a second enzyme and hydrogen source like formate/ formate dehydrogenase or glucose/ glucose dehydrogenase. Another common way is to add a sacrificial cheap alcohol like isopropanol which is oxidised to acetone to regenerate NAD(P)+ to NAD(P)H.
Key references
Kratzer, R.; Woodley, J. M.; Nidetzky, B. Rules for biocatalyst and reaction engineering to implement effective, NAD(P)H-dependent, whole cell bioreductions. Biotechnol. Adv. 2015, 33, 1641–1652.
Thai, Y.-C.; Szekrenyi, A.; Qi, Y.; Black, G. W.; Charnock, S. J.; Fessner, W.-D. Fluorogenic kinetic assay for high-throughput discovery of stereoselective ketoreductases relevant to pharmaceutical synthesis. BioOrg. Med. Chem. 2017, 26, 1320–1326.
Ni, Y.; Xu, J.-H. Biocatalytic ketone reduction: A green and efficient access to enantiopure alcohols. Biotechnol. Adv. 2012, 30, 1279–1288.
Rowan, A. S.; Moody, T. S.; Howard, R. M.; Underwood, T. J.; Miskelly, I. R.; He, Y.; Wang, B. Preparative access to medicinal chemistry related chiral alcohols using carbonyl reductase technology. Tet. Asymmetry 2013, 24,1369–1381.
Huisman, G. W.; Liang, J.; Krebber, A. Practical chiral alcohol manufacture using ketoreductases. Curr. Opin. Chem. Biol. 2009, 14, 1–8.
Kaluzna, I. A.; Rozzell, J. D.; Kambourakis, S. Ketoreductases: Stereoselective catalysts for the facile synthesis of chiral alcohols. Tet. Asymmetry 2005, 16, 3682–3689.
Calvin, S. J.; Mangan, D.; Miskelly, I.; Moody, T. S.; Stevenson, P. J. Overcoming Equilibrium Issues with Carbonyl Reductase Enzymes. Org. Process Res. Dev. 2012, 16, 82−86.
Modukuru, N. K.; Sukumaran, J.; Collier, S. J.; Chan, A. S.; Gohel, A.; Huisman, G. W.; Keledjian, R.; Narayanaswamy, K.; Novick, S. J.; Palanivel, S. M.; et al. Development of a Practical, Biocatalytic Reduction for the Manufacture of (S)-Licarbazepine Using an Evolved Ketoreductase. Org. Process Res. Dev. 2014, 18, 810−815.
Zou, J.; Ni, G.; Tang, J.; Yu, J.; Jiang, L.; Ju D.; Zhang, F.; Chen, S. Asymmetric Synthesis of Florfenicol by Dynamic Reductive Kinetic Resolution with Ketoreductases. Eur. J. Chem. 2018, 36, 5044-5053.
Li, H.; Moncecchi, J.; Truppo, M. D. Development of an Immobilized Ketoreductase for Enzymatic (R)‑1-(3,5-Bis(trifluoromethyl)phenyl)ethanol Production. Org. Process Res. Dev. 2015, 19, 695−700.
Magnus, N. A.; Coffey, D. S.; DeBaillie, A. C.; Jones, C. A.; Kaluzna, I. A.; Kambourakis, S.; Pu, Y. J.; Wang, L.; Wepsiec, J. P. Diarylketone Ketoreductase Screen and Synthesis Demonstration to Access mGlu2 Receptor Potentiators. Org. Process Res. Dev. 2011, 15, 1377–1381.
Dascier, D.; Kambourakis, S.; Hua, L.; Rozzell, J. D.; Stewart, J. D. Influence of Cofactor Regeneration Strategies on Preparative-Scale, Asymmetric Carbonyl Reductions by Engineered Escherichia coli. Org. Process Res. Dev. 2014, 18, 793–800.
Kratzer, R.; Woodley, J. M.; Nidetzky, B. Rules for biocatalyst and reaction engineering to implement effective, NAD(P)H-dependent, whole cell bioreductions. Biotechnol. Adv. 2015, 33 (8), 1641–1652.
Relevant scale up examples
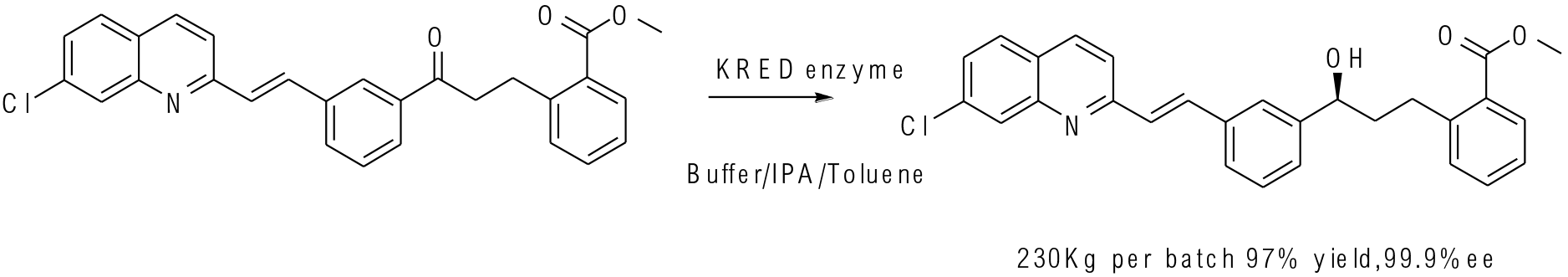
Org. Process Res. Dev. 2010, 14, 193−198
230 kg scale
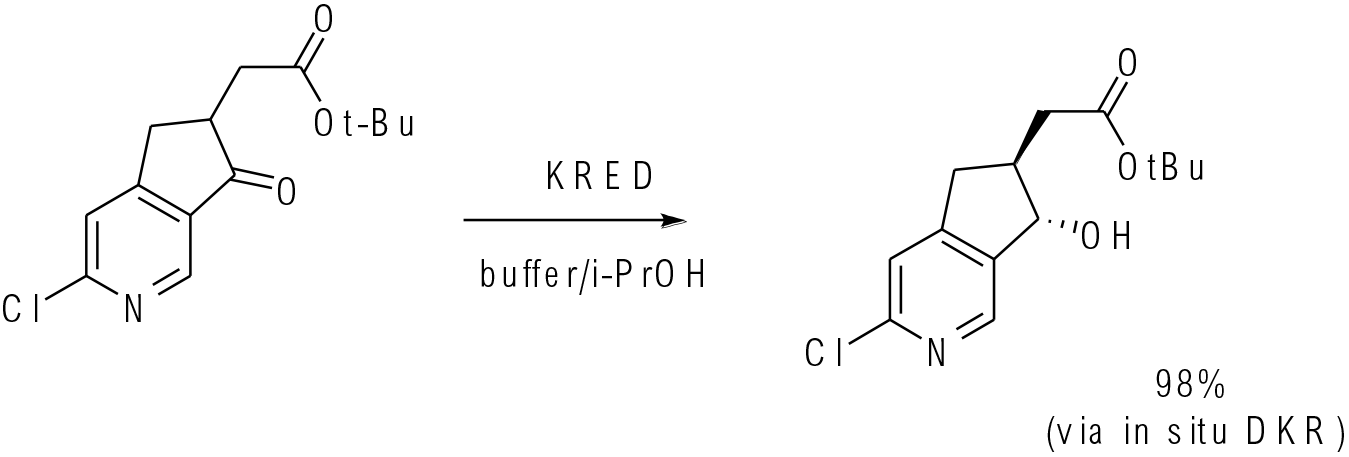
Org. Lett. 2016, 18, 5888−5891
2 g scale
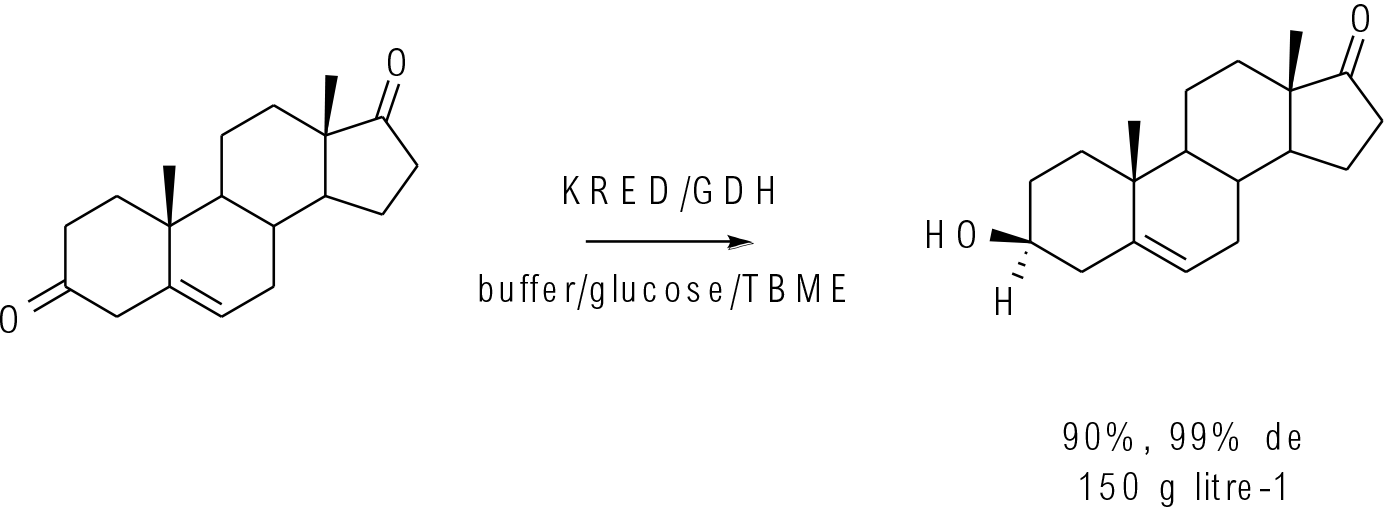
Org. Process Res. Dev. 2016, 20, 1520−1528
3 g scale
Green Review
-
Atom efficiency (by-products, molecular weight)
Catalytic technology—but enzyme loading should be optimized and steps using stoichiometric reagents like cofactor recycle should be optimized to avoid unnecessary excess of reagents.
- Safety Concerns
Generally considered a safe technology to scale-up. Issues—enzymes/proteins can be sensitizers by inhalation, and some by mode of action can be skin irritants (proteases). If viable GMO cells are used, local regulations relating to use of GMOs need to be followed.
- Toxicity and environmental/aquatic impact
No real concerns—enzymes are non-toxic and readily biodegradable. If viable GMO cells are used, these need to be passivated before discharge into the environment.
- Cost, availability & sustainable feedstocks
Enzymes are produced from natural sustainable feedstocks.
- Sustainable implications
Enzymes are made via fermentation; cloned and recombinant enzymes would be at no risk from depletion. Enzymes break down in the environment and the constituent amino acids are recycled in nature. The use of over expressed recombinant enzymes is generally much better than natural enzymes on a life cycle impact basis.